Registration for NGSchool 2022: Machine Learning in Computational Biology is now closed. Thank you to all who took time to answer the questions and solve the problems in the registration form. Here we publish the solutions.
Coding questions
The following questions were meant to ensure that the applicants have basic coding skills and will be able to follow the practical part of the school. We accepted solutions in any programming language, but we recommend using Python or R, as the practical part of the NGSchool program will be conducted using those. The coding solutions provided below are in R.
C1 - Fibonacci sequence
The Fibonacci sequence starts with 0 and 1 (positions 1 and 2). Each next element is a sum of two preceding elements. What is the difference between elements at positions 55th and 45th multiplied by the element at position 5th?
Solution: 256698487617
# create empty vector
fl <- vector("integer", 100L)
fl[1] <- 0
fl[2] <- 1
# create Fibonacci sequence
for (i in 3:100) {
fl[i] <- fl[i-1] + fl[i-2]
}
(fl[55] - fl[45]) * fl[5] # 256698487617C2 - Protein sequence
Here you can find the aminoacid sequence of BRCA2 (each letter represents one amino acid). Which amino acids are the most and the least frequent ones in this sequence and how many times they are present?
A) Least frequent: threonine (T, 30 times), most frequent: serine (S, 318 times)
B) Least frequent: tyrosine (Y, 79 times), most frequent: lysine (K, 322 times)
C) Least frequent: cysteine (C, 76 times), most frequent: selenocysteine (U, 413 times)
D) Least frequent: tryptophan (W, 20 times), most frequent: serine (S, 381 times)
E) Least frequent: phenylalanine (F, 18 times), most frequent: selenocysteine (U, 238 times)
Solution: D)
brca2 <- "MPIGSKERPTFFEIFKTRCNKADLGPISLNWFEELSSEAPPYNSEPAEESEHKNNNYEPNLFKTPQRKPSYNQLASTPIIFKEQGLTLPLYQSPVKELDKFKLDLGRNVPNSRHKSLRTVKTKMDQADDVSCPLLNSCLSESPVVLQCTHVTPQRDKSVVCGSLFHTPKFVKGRQTPKHISESLGAEVDPDMSWSSSLATPPTLSSTVLIVRNEEASETVFPHDTTANVKSYFSNHDESLKKNDRFIASVTDSENTNQREAASHGFGKTSGNSFKVNSCKDHIGKSMPNVLEDEVYETVVDTSEEDSFSLCFSKCRTKNLQKVRTSKTRKKIFHEANADECEKSKNQVKEKYSFVSEVEPNDTDPLDSNVANQKPFESGSDKISKEVVPSLACEWSQLTLSGLNGAQMEKIPLLHISSCDQNISEKDLLDTENKRKKDFLTSENSLPRISSLPKSEKPLNEETVVNKRDEEQHLESHTDCILAVKQAISGTSPVASSFQGIKKSIFRIRESPKETFNASFSGHMTDPNFKKETEASESGLEIHTVCSQKEDSLCPNLIDNGSWPATTTQNSVALKNAGLISTLKKKTNKFIYAIHDETSYKGKKIPKDQKSELINCSAQFEANAFEAPLTFANADSGLLHSSVKRSCSQNDSEEPTLSLTSSFGTILRKCSRNETCSNNTVISQDLDYKEAKCNKEKLQLFITPEADSLSCLQEGQCENDPKSKKVSDIKEEVLAAACHPVQHSKVEYSDTDFQSQKSLLYDHENASTLILTPTSKDVLSNLVMISRGKESYKMSDKLKGNNYESDVELTKNIPMEKNQDVCALNENYKNVELLPPEKYMRVASPSRKVQFNQNTNLRVIQKNQEETTSISKITVNPDSEELFSDNENNFVFQVANERNNLALGNTKELHETDLTCVNEPIFKNSTMVLYGDTGDKQATQVSIKKDLVYVLAEENKNSVKQHIKMTLGQDLKSDISLNIDKIPEKNNDYMNKWAGLLGPISNHSFGGSFRTASNKEIKLSEHNIKKSKMFFKDIEEQYPTSLACVEIVNTLALDNQKKLSKPQSINTVSAHLQSSVVVSDCKNSHITPQMLFSKQDFNSNHNLTPSQKAEITELSTILEESGSQFEFTQFRKPSYILQKSTFEVPENQMTILKTTSEECRDADLHVIMNAPSIGQVDSSKQFEGTVEIKRKFAGLLKNDCNKSASGYLTDENEVGFRGFYSAHGTKLNVSTEALQKAVKLFSDIENISEETSAEVHPISLSSSKCHDSVVSMFKIENHNDKTVSEKNNKCQLILQNNIEMTTGTFVEEITENYKRNTENEDNKYTAASRNSHNLEFDGSDSSKNDTVCIHKDETDLLFTDQHNICLKLSGQFMKEGNTQIKEDLSDLTFLEVAKAQEACHGNTSNKEQLTATKTEQNIKDFETSDTFFQTASGKNISVAKESFNKIVNFFDQKPEELHNFSLNSELHSDIRKNKMDILSYEETDIVKHKILKESVPVGTGNQLVTFQGQPERDEKIKEPTLLGFHTASGKKVKIAKESLDKVKNLFDEKEQGTSEITSFSHQWAKTLKYREACKDLELACETIEITAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNLCRQTENLKTSKSIFLKVKVHENVEKETAKSPATCYTNQSPYSVIENSALAFYTSCSRKTSVSQTSLLEAKKWLREGIFDGQPERINTADYVGNYLYENNSNSTIAENDKNHLSEKQDTYLSNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVKDANAYPQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIKKVKDIFTDSFSKVIKENNENKSKICQTKIMAGCYEALDDSEDILHNSLDNDECSTHSHKVFADIQSEEILQHNQNMSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFSTASGKSVQVSDASLQNARQVFSEIEDSTKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLISQKGFSYNVVNSSAFSGFSTASGKQVSILESSLHKVKGVLEEFDLIRTEHSLHYSPTSRQNVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSENNHSIKVSPYLSQFQQDKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTYSKDSENYFETEAVEIAKAFMEDDELTDSKLPSHATHSLFTCPENEEMVLSNSRIGKRRGEPLILVGEPSIKRNLLNEFDRIIENQEKSLKASKSTPDGTIKDRRLFMHHVSLEPITCVPFRTTKERQEIQNPNFTAPGQEFLSKSHLYEHLTLEKSSSNLAVSGHPFYQVSATRNEKMRHLITTGRPTKVFVPPFKTKSHFHRVEQCVRNINLEENRQKQNIDGHGSDDSKNKINDNEIHQFNKNNSNQAVAVTFTKCEEEPLDLITSLQNARDIQDMRIKKKQRQRVFPQPGSLYLAKTSTLPRISLKAAVGGQVPSACSHKQLYTYGVSKHCIKINSKNAESFQFHTEDYFGKESLWTGKGIQLADGGWLIPSNDGKAGKEEFYRALCDTPGVDPKLISRIWVYNHYRWIIWKLAAMECAFPKEFANRCLSPERVLLQLKYRYDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANISETSSNKTSSADTQKVAIIELTDGWYAVKAQLDPPLLAVLKNGRLTVGQKIILHGAELVGSPDACTPLEAPESLMLKISANSTRPARWYTKLGFFPDPRPFPLPLSSLFSDGGNVGCVDVIIQRAYPIQWMEKTSSGLYIFRNEREEEKEAAKYVEAQQKRLEALFTKIQEEFEEHEENTTKPYLPSRALTRQQVRALQDGAELYEAVKNAADPAYLEGYFSEEQLRALNNHRQMLNDKKQAQIQLEIRKAMESAEQKEQGLSRDVTTVWKLRIVSYSKKEKDSVILSIWRPSSDLYSLLTEGKRYRIYHLATSKSKSKSERANIQLAATKKTQYQQLPVSDEILFQIYQPREPLHFSKFLDPDFQPSCSEVDLIGFVVSVVKKTGLAPFVYLSDECYNLLAIKFWIDLNEDIIKPHMLIAASNLQWRPESKSGLLTLFAGDFSVFSASPKEGHFQETFNKMKNTVENIDILCNEAENKLMHILHANDPKWSTPTKDCTSGPYTAQIIPGTGNKLLMSSPNCEIYYQSPLSLCMAKRKSVSTPVSAQMTSKSCKGEKEIDDQKNCKKRRALDFLSRLPLPPPVSPICTFVSPAAQKAFQPPRSCGTKYETPIKKKELNSPQMTPFKKFNEISLLESNSIADEELALINTQALLSGSTGEKQFISVSESTRTAPTSSEDYLRLKRRCTTSLIKEQESSQASTEECEKNKQDTITTKKYI"
aa <- strsplit(brca2,"")[[1]]
sort(table(aa))
# W M C Y H R G F P Q D A V I T N L E K S
# 20 45 76 79 83 110 122 136 149 154 171 174 185 187 220 230 281 293 322 381 C3 - Gene mutations
Here you will find 2 nucleotide sequences. They come from the MT-CYB gene that is quite special, because it consists of only 1 exon, thus the whole sequence is a coding region. First sequence is a reference one and the second is one with some mutation. Which of the mutations is present in this sequence and what it results in?
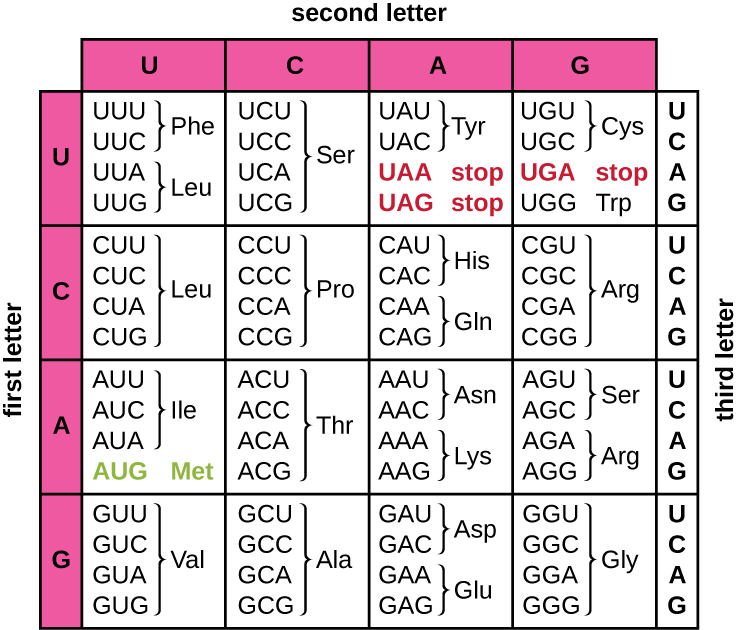
A) G to A at position 752, resulting in change from glycine to asparagine
B) G to A at position 869, resulting in change from glycine to threonine
C) G to A at position 751, resulting in change from glycine to serine
D) C to A at position 496, resulting in change from glycine to threonine
E) G to A at position 826, resulting in change from phenylalanine to leucine
Solution: C)
wt <- "ATGACCCCAATACGCAAAACTAACCCCCTAATAAAATTAATTAACCACTCATTCATCGACCTCCCCACCCCATCCAACATCTCCGCATGATGAAACTTCGGCTCACTCCTTGGCGCCTGCCTGATCCTCCAAATCACCACAGGACTATTCCTAGCCATGCACTACTCACCAGACGCCTCAACCGCCTTTTCATCAATCGCCCACATCACTCGAGACGTAAATTATGGCTGAATCATCCGCTACCTTCACGCCAATGGCGCCTCAATATTCTTTATCTGCCTCTTCCTACACATCGGGCGAGGCCTATATTACGGATCATTTCTCTACTCAGAAACCTGAAACATCGGCATTATCCTCCTGCTTGCAACTATAGCAACAGCCTTCATAGGCTATGTCCTCCCGTGAGGCCAAATATCATTCTGAGGGGCCACAGTAATTACAAACTTACTATCCGCCATCCCATACATTGGGACAGACCTAGTTCAATGAATCTGAGGAGGCTACTCAGTAGACAGTCCCACCCTCACACGATTCTTTACCTTTCACTTCATCTTGCCCTTCATTATTGCAGCCCTAGCAACACTCCACCTCCTATTCTTGCACGAAACGGGATCAAACAACCCCCTAGGAATCACCTCCCATTCCGATAAAATCACCTTCCACCCTTACTACACAATCAAAGACGCCCTCGGCTTACTTCTCTTCCTTCTCTCCTTAATGACATTAACACTATTCTCACCAGACCTCCTAGGCGACCCAGACAATTATACCCTAGCCAACCCCTTAAACACCCCTCCCCACATCAAGCCCGAATGATATTTCCTATTCGCCTACACAATTCTCCGATCCGTCCCTAACAAACTAGGAGGCGTCCTTGCCCTATTACTATCCATCCTCATCCTAGCAATAATCCCCATCCTCCATATATCCAAACAACAAAGCATAATATTTCGCCCACTAAGCCAATCACTTTATTGACTCCTAGCCGCAGACCTCCTCATTCTAACCTGAATCGGAGGACAACCAGTAAGCTACCCTTTTACCATCATTGGACAAGTAGCATCCGTACTATACTTCACAACAATCCTAATCCTAATACCAACTATCTCCCTAATTGAAAACAAAATACTCAAATGGGCCT"
mut <- "ATGACCCCAATACGCAAAACTAACCCCCTAATAAAATTAATTAACCACTCATTCATCGACCTCCCCACCCCATCCAACATCTCCGCATGATGAAACTTCGGCTCACTCCTTGGCGCCTGCCTGATCCTCCAAATCACCACAGGACTATTCCTAGCCATGCACTACTCACCAGACGCCTCAACCGCCTTTTCATCAATCGCCCACATCACTCGAGACGTAAATTATGGCTGAATCATCCGCTACCTTCACGCCAATGGCGCCTCAATATTCTTTATCTGCCTCTTCCTACACATCGGGCGAGGCCTATATTACGGATCATTTCTCTACTCAGAAACCTGAAACATCGGCATTATCCTCCTGCTTGCAACTATAGCAACAGCCTTCATAGGCTATGTCCTCCCGTGAGGCCAAATATCATTCTGAGGGGCCACAGTAATTACAAACTTACTATCCGCCATCCCATACATTGGGACAGACCTAGTTCAATGAATCTGAAGAGGCTACTCAGTAGACAGTCCCACCCTCACACGATTCTTTACCTTTCACTTCATCTTGCCCTTCATTATTGCAGCCCTAGCAACACTCCACCTCCTATTCTTGCACGAAACGGGATCAAACAACCCCCTAGGAATCACCTCCCATTCCGATAAAATCACCTTCCACCCTTACTACACAATCAAAGACGCCCTCGGCTTACTTCTCTTCCTTCTCTCCTTAATGACATTAACACTATTCTCACCAGACCTCCTAAGCGACCCAGACAATTATACCCTAGCCAACCCCTTAAACACCCCTCCCCACATCAAGCCCGAATGATATTTCCTATTCGCCTACACAATTCTCCGATCCGTCCCTAACAAACTAGGAGACGTCCTTGCCCTATTACTATCCATCCTCATCCTAGCAATAATCCCCATCCTCCATATATCCAAACAACAAAGCATAATATTTCGCCCACTAAGCCAATCACTTTATTGACTCCTAGCCGCAGACCTCCTCATTCTAACCTGAATCGGAGGACAACCAGTAAGCTACCCTTTTACCATCATTGGACAAGTAGCATCCGTACTATACTTCACAACAATCCTAATCCTAATACCAACTATCTCCCTAATTGAAAACAAAATACTCAAATGGGCCT"
wt_aa <- strsplit(wt,"")[[1]]
mut_aa <- strsplit(mut,"")[[1]]
pos_mut <- which(wt_aa != mut_aa)
pos_mut # 496 751 869
# get bases that change
sapply(pos_mut, function(pos) c(wt_aa[pos], mut_aa[pos]))
# [,1] [,2] [,3]
# [1,] "G" "G" "G"
# [2,] "A" "A" "A"
# get codons that change
sapply(pos_mut, function(pos) {
codon_start = pos%/%3 * 3
codon_end = codon_start + 2
c(
paste0(wt_aa[codon_start:codon_end],collapse=""),
paste0(mut_aa[codon_start:codon_end],collapse="")
)
})
[,1] [,2] [,3]
[1,] "AGG" "AGG" "AGG"
[2,] "AAG" "AAG" "AGA"
# AGG -> AAG: Arg -> Lys
# AGG -> AAG: Arg -> Lys
# AGG -> AGA: Arg -> ArgC4 to C6 - scRNA-seq database
Single-cell RNA sequencing (scRNA-seq) is arguably the most dramatically growing technology in both scale and use today. A curated database of scRNA-seq studies is available at https://www.nxn.se/single-cell-studies. Answer the following questions using the snapshot of data from github.
C4: How many studies report data from more than one organism?
Solution: 146
require(data.table)
# load data
url <- "https://raw.githubusercontent.com/NGSchoolEU/ngs22_registration_form/1cc647a3733e2c8a21b47aa497b4ca8c42457aa8/data/single-cell-studies.tsv"
scs <- fread(url)
# select the studies that report data from more than one organism
dups <- scs[grep(",", Organism)]
# notice that one entry appears twice
dups[,.N,.(Authors,Journal,DOI)][order(-N)]
# count only unique entries
length(unique(dups$DOI)) # 146Half of the applicants answered this wrongly only because of not removing one duplicated study entry. Only 4% of applicants got it right.
C5: After excluding the studies that report data from more than one organism, for which organism there are the most published studies, and how many?
Solution: Mouse, 632
# remove studies reporting data for more than one organism
scst <- scs[!DOI %in% dups$DOI]
# select nonduplicated entries
sc_studies <- scst[,.N,.(Authors,Journal,DOI,Organism)][order(-N)]
# summarize number of studies by organism
sc_studies[,.N,Organism][Organism!=""][order(-N)][1] # Mouse 632So much as 56% of applicants got this right - higher than C4 or C6 primarily because there are no duplicated study entries for mouse.
# same result even if we don't remove duplicated entry
scst[,.N,Organism][Organism!=""][order(-N)][1] # Mouse 632C6: After excluding the studies that report data from more than one organism, for which organism there are most reported cells, and how many?
Solution: Human, 36084826
# parse number of cells which is stored as character
sc <- scst[!is.na(`Reported cells total`)][,Cells:=as.integer(stringr::str_remove_all(`Reported cells total`,","))]
# select nonduplicated entries
sc_cells <- sc[,.N,.(Authors,Journal,DOI,Organism,Cells)][order(-N)]
# summarize number of cells by organism
sc_cells[,.(N=sum(Cells,na.rm=TRUE)),Organism][Organism!=""][order(-N)][1] # Human 36084826Most wrong answers here include not parsing the cell number correctly and not removing the same duplicated study as in C4.
# without parsing cell number stored as character
scst[,.(N=sum(as.integer(`Reported cells total`),na.rm=TRUE)),Organism][order(-N)][1] # Mouse 48131
# without removing duplicated entries
sc[,.(N=sum(Cells,na.rm=TRUE)),Organism][Organism!=""][order(-N)][1] # Human 36199222Statistics and probability
The following questions were supposed to make sure that you are familiar with basic concepts in probability and statistics.
SP1 and SP2
Celiac disease is an autoimmune disease that affects 1 in 100 worldwide. For people that have this disease, the ingestion of gluten causes serious damage in the small intestine and, if untreated, it can lead to development of other long-term health conditions. Celiac disease is hereditary - people with a first-degree relative with celiac disease (parent, child, sibling) have a 1 in 10 risk of developing celiac disease. A new diagnostic test has appeared on the market that claims to have a 95% accuracy in early detection of celiac disease.
SP1: What is the probability that a person who gets a positive result actually has Celiac disease, if no other members in their family have it?
Solution: 0.16
SP2: What is the probability that a person who gets a positive result actually has Celiac disease, if they also have a first-degree relative diagnosed with it?
Solution: 0.68
Explanation:
An accuracy of a test is measured as the number of cases it correctly detects as positives (true positive, TP), plus the number of controls it correctly reports as negative (true negative, TN), out of the total number of samples tested (including TP, TN, and false positives FP, and false negatives FN).
Take a hypothetical scenario for SP1. If we tested 10,000 people, 100 of which have Celiac disease, a test with 0.95 accuracy would report 0.95 x 100 = 95 TP and 0.95 x 9,900 = 9,405 TN. It would also wrongly report 0.05 x 100 = 5 FN (people with disease who would get a negative test result), and 0.05 x 9,900 = 495 FP. Fraction of TP out of all the positives reported by the test is 95 / (95 + 495) = 0.16.
In the SP2 scenario, the prevalence of disease is 10 times higher, so 1,000 out of 10,000 cases would be positive. Now the test with 95% accuracy would report 0.95 x 1,000 = 950 TP and 0.95 x 9,000 = 8,550 TN. It would also wrongly report 0.05 x 1,000 = 50 FN and 0.05 x 9,000 = 450 FP. Fraction of TP out of all the positives reported by the test now is 950 / (950 + 450) = 0.68.
Note:
The explained solutions assume that the reported accuracy of the test is the same for both groups of people (healthy and patients). As this was not stated explicitly, we will accept as correct answers those that said there was not enough information to calculate this.
SP3
In a study that looked at the effect of substance A on the global gene expression levels of 20 thousand genes in 10 cell line samples, authors report significantly reduced and increased expression for 1287 and 446 genes, respectively (t-test, p value<0.05). Which of the following problems occurs in this analysis?
A) T-test not applicable because of sample size
B) Imbalanced classes
C) No adjustment for multiple comparisons
D) Comparison with another substance
E) No correction for ethnicity of sample donors
Solution: C)
SP5
In the same experiment, their favourite gene X is found to be differentially expressed with a large absolute fold change and p value of 0.01. This means that…
A) there is 1% probability that gene X with this or larger absolute fold change is not differentially expressed
B) there is 1% probability that gene X would have this or larger absolute fold change if it was not differentially expressed
C) the confidence that gene X is differentially expressed is 99%
D) the significance of the differential expression of the gene X is 99%
E) the variability of gene X expression differs significantly between groups and is higher than in 99% of other genes
Solution: B)
SP6
Select all relationships, which should be modelled with linear regression (multiple choice)
A) protein concentration dependance on gene expression
B) birth weight dependance on duration of pregnancy in weeks
C) Alzheimer’s disease occurrence depending on gender and age
D) life expectancy in cancer patients depending on TNM cancer stage
E) A-F grade received on a test with respect to meat consumption (in kg) in the last month
F) fever occurrence depending on recent bacterial infection
Solution: A), B), D)
Biological study design and interpretation
The last set of question checked that the applicants understand basics of biological study design and can critically interpret presented results.
R1
In the picture below you can find a figure and fragment of its description from the paper on the role of metformin in survival of pancreatic cancer patients. The paper describes a metaanalysis that is a statistical compilation of many similar studies on the specific topic. This metaanalysis included randomised clinical trials and observational studies done in many clinics.
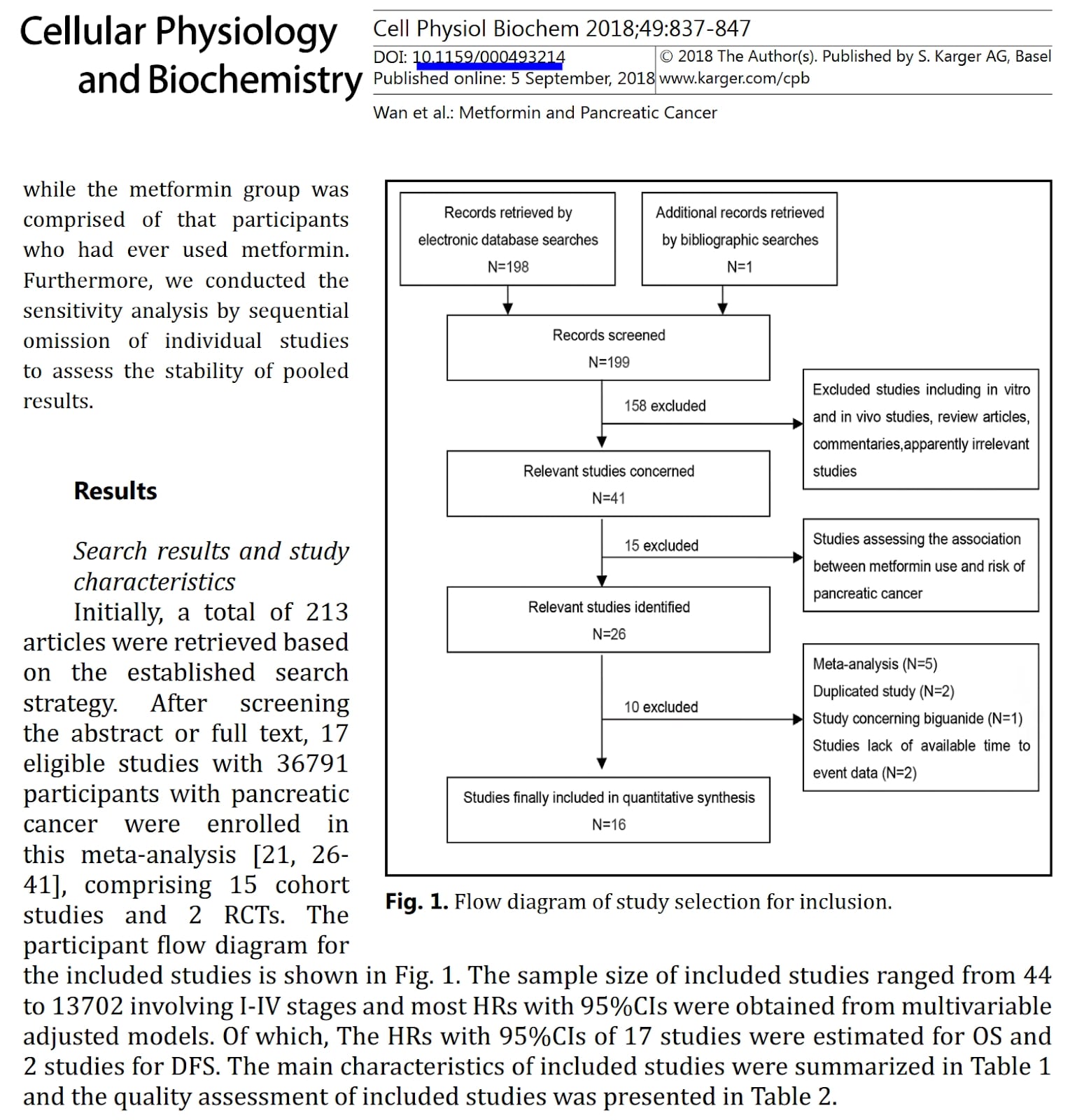
What problem with this study can you identify based on the provided snippet?
A) Commentaries should not be excluded
B) Sample sizes from different studies are too variable to combine all those studies together
C) Numbers of articles are inconsistent between text and figure
D) Literature review was conducted in wrong order
E) Cell line studies should not be excluded
Solution: C)
R2
In the picture you can find a figure with its caption from the paper paper on the mutational landscape of 12 types of cancers . Analysed types of cancers are identified by abbreviations at the bottom of panel a.
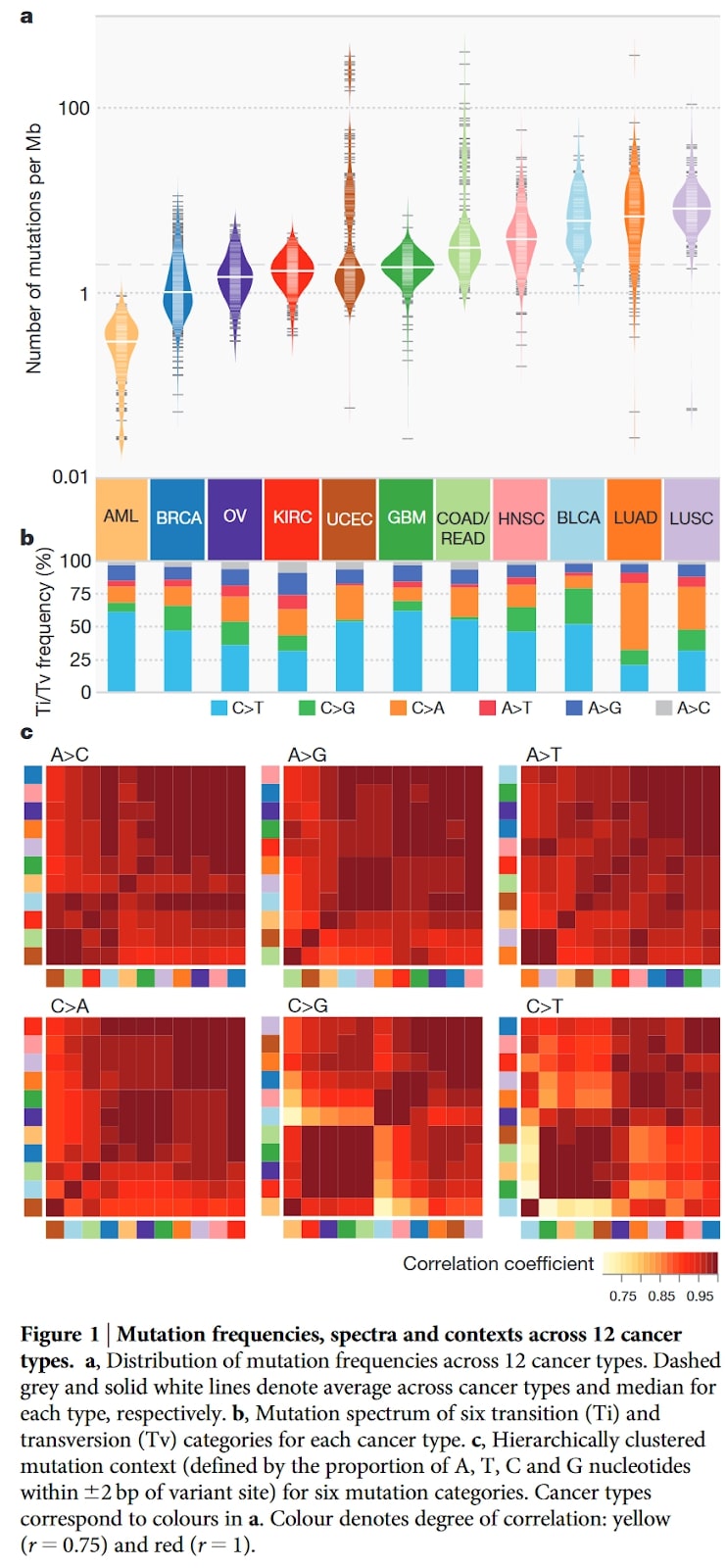
Which of the following is true about this figure?
A) Mutations in ovarian cancer (OV) are more frequent than in bladder cancer (BLCA)
B) Changes from A to T are the most frequent ones in majority of cancers
C) Glioblastomas (GMB) are a cancer type with the greatest number of outliers
D) Proportion of mutations (panel b) are very similar in lung adenocarcinoma (LUAD) and breast adenocarcinoma (BRCA)
E) Lung cancers (LUAD and LUSC) have more C to A mutations than other cancers
Solution: E)
R3
In the picture you can see a figure from the paper on acute lymphoblastic leukaemia in children. The figure shows how the survival rate of the patients enrolled in clinical trials changed from 1968 to 2009.
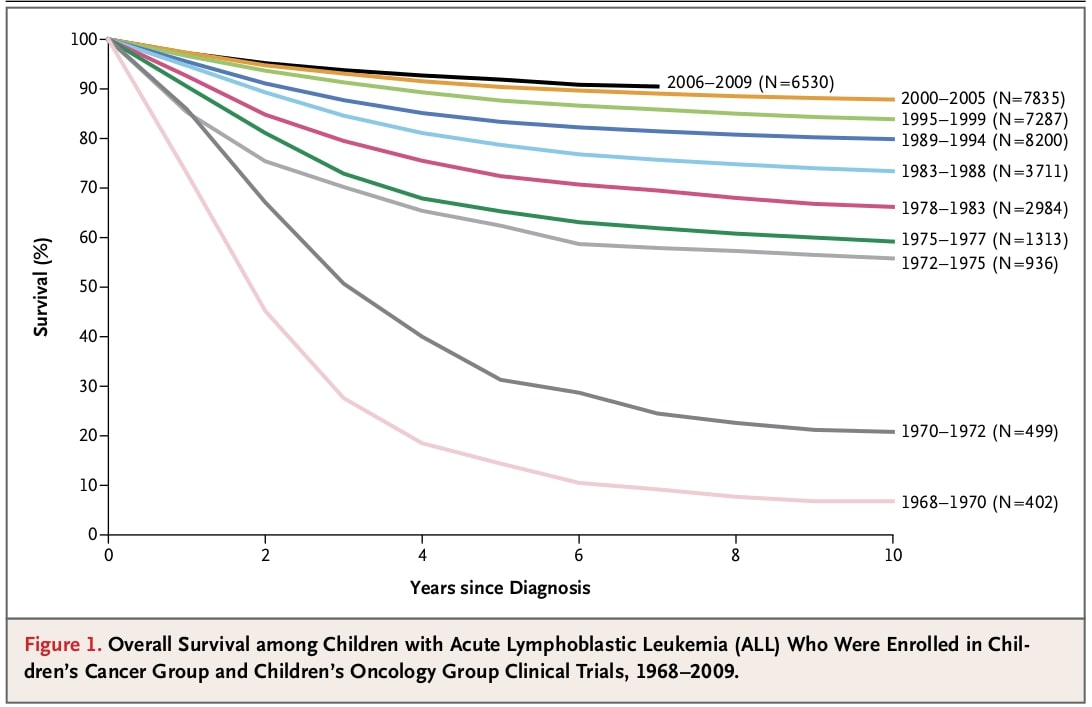
Select all answers that correctly describe this plot.
A) None of the children who were enrolled between 1968-70 survived more than 8 years.
B) From 1972 on median survival is longer than 10 years.
C) More than 80% of children enrolled in trials in the 21st century survive more than 5 years.
D) Number of children enrolled in trials from 1968 to 1994 is higher than in the period from 1995 to 2009.
E) 5-years survival constantly improved in the analysed period.
Solution: B), C), E)
Conclusion
We hope you enjoyed the problems in this year's NGSchool application form. Stay tuned for the announcement of the application results in the beginning of July!
Also check out the program for NGSymposium2022 which will take place immediately after the summer school. There is still time to submit abstract for a chance to present your work at an international conference in Warsaw.

